MGIZA++ 4.0.1 Crack Free For PC
- ryoraflingcingling
- May 20, 2022
- 4 min read
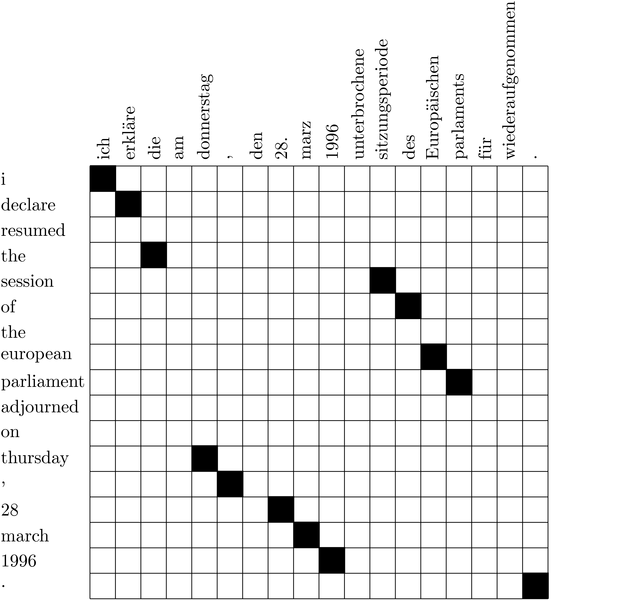
MGIZA++ Free Download 2022 MGIZA++ is a multi-threaded, but also very simple and fast word alignment tool. If you are looking for an open source, fast and easy to use solution to word alignment, the MGIZA++ package is the one to consider. MGIZA++ features a one-line command line and no difficult or confusing parameters. You can edit the alignments at the point in time and save your document using a generic file format. This tool is open source and distributed under the GNU General Public License (GPL) v3. MGIZA++ home page: Requirements: GNU/Linux only. LGPL License: This software is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. You can read more about the LGPL license here: Support and Help: You can always contact us via the support forum: You can also contact us via email. Please specify the version of MGIZA++ you are using. Email: mzmike@gmail.com Version 1.5.0 (2020-03-03) Known Bugs: * When applying the alignment on documents which have a single transcript, the result is sometimes ignored. * When applying the alignment on documents which have multiple transcripts, the result is sometimes ignored. * There is no difference between transcript_id's when setting the ignore_replicates parameter. * When applying the alignment on documents which have a single transcript, the result is sometimes ignored. * When applying the alignment on documents which have multiple transcripts, the result is sometimes ignored. * When applying the alignment on documents which have multiple transcripts and multiple transcripts have the same transcript_id, the result is sometimes ignored. * When applying the alignment on documents which have multiple transcripts, the result is sometimes ignored. * When applying the alignment on documents which have multiple transcripts and one transcript has the same transcript_id and the other transcripts have transcript_ids which are not the same, the result is sometimes MGIZA++ Crack + PC/Windows Latest This module integrates MGIZA++ Crack Keygen Word Alignment Tool with Joblib. MGIZA++ Torrent Download is used for alignment, which analyzes the alignment between a pair of word sequences. User can simply choose several criteria and MGIZA++ will return the most similar word sequences. For example, user can input two sequences, and then use a pre-defined similarity score. Or user can input the real alignment file. To use this module, you need a compiled MGIZA++(or even a pre-compiled version) Input: For the input data, it is required to provide an alignment file(One alignment for two sequences). For the other input, it is required to provide a file of sequence that match your chosen criteria. In the input data, user can input: 1. A file of sequence pair(s) and specify the scores and cutoffs for your selected criteria. 2. A file of sequence and specify the scores and cutoffs for your selected criteria. Note: The File must be in FASTA format. Sequence must be sorted. Output: The output of MGIZA++ is a file of word sequence pair(s). Example: mriza ++ -h giza++ -o filename -n 1 -s 0 -p 0 -i giza++.in -o giza++.out -t In the above example, it means you want to calculate the similarity score and distance between sequence "pair1" and sequence "pair2" And you can choose different criterias for pair1 and pair2 You need to specify the score and cutoff for each criterias, "0" means you don't care about it. Use: To calculate the score, you need a compiled MGIZA++(or even a pre-compiled version) For a pre-compiled version, please refer to: For more information about MGIZA++, please refer to For more information about Joblib, please refer to Camden's New Look Camden's New Look is a collection of printmaker's works by the English printmaker Peter Lely (1618-1680). It was one of the first substantial collections of prints by an English artist 1a423ce670 MGIZA++ GIZA++ is a program written in C++ that aims to replace the tedious task of RNA sequence alignment, an essential step in studying RNA function. By default, GIZA++ will align multiple input sequences, aligning each sequence to every other, against a single input sequence or database. You can use the GIZA++ to create alignments with subsets of input sequences and with subsets of sequences from a specific organism. You can output your results in various formats and use GIZA++ to align not just RNA sequences but other protein and DNA sequences as well. THE DIAGNOSTIC AND STATISTICAL TOOLS in MGIZA++ 1. CAST: Clustering based on Similarity and Arrangement of Transcripts 2. T-COFFE: The Transcript Coding Fuzzy Extractor 3. PICARD: The Transcript Coding Identification and Alignment Coverage 4. NBI: The No Better Explanation Analysis Tool 5. CDD: Conserved Domains Database 6. InterProScan: The InterProScan 7. Blastall: The Blastall 8. BlastallD: The BlastallD 9. RATS: RNA Transcripts in SWISS-PROT MGIZA++ Usage: Pre-Installation Guide: Using MGIZA++ After Installation Guide: Using MGIZA++ How to use MGIZA++ (click "README.TXT" on the MGIZA++ Package to get a short introduction of MGIZA++) Installing the Downloaded Package Installing the CURRENT Version in Windows: 1. Extract the MGIZA++ package 2. Locate the MGIZA++ folder 3. Create a shortcut to the MGIZA++ folder 4. Install the shortcut to the Start Menu Installing the Downloaded Package in Mac OS: 1. Move the MGIZA++ folder to the Library folder 2. Install the shortcut to the Start Menu Please Note: This tool is designed to run on Windows or Mac OS. It is not compatible with Linux. General Usage: 1. Download the MGIZA++: (1) For Windows The latest version of the MGIZA++ is located at: (2) For Mac OS The latest version of the MGIZA++ is located at: 2. Extract the MGIZ What's New in the MGIZA ? System Requirements For MGIZA : Minimum: OS: Windows 7 or later (64-bit) CPU: Intel Core 2 Duo or later Memory: 2 GB RAM Graphics: DirectX 10 or later DirectX: Version 9.0c Network: Broadband internet connection Storage: 600 MB available space Additional Notes: Internet connection required to download and play the game Recommended: CPU: Intel Core i3 or later Memory: 4 GB RAM
Related links:

![Download Lego City Pc Game With Crack Torrent [CRACKED]](https://static.wixstatic.com/media/698717_4f3caa50635b4c15a45bbff01a811d27~mv2.jpeg/v1/fill/w_720,h_450,al_c,q_80,enc_avif,quality_auto/698717_4f3caa50635b4c15a45bbff01a811d27~mv2.jpeg)

Comments